Abstract
Acute myeloid leukemia (AML) is a clinically and biologically heterogeneous disease. Traditionally, cytogenetic analysis has been the backbone for prognostication and treatment decisions. Outcomes vary between age groups with older adults generally having a poorer prognosis. Next Generation Sequencing (NGS) has expedited the discovery of novel genetic lesions in AML to better predict response to intensive chemotherapy and overall survival (OS).
The aims of our study were to describe the genetic profile of older adults with AML and to determine its impact on treatment response and survival.
We included all new patients with a diagnosis of AML (≥20% blasts in peripheral blood or bone marrow; acute promyelocytic leukemia and myeloid sarcoma were excluded), treated at Princess Margaret Cancer Centre between February 2015 and August 2017.
NGS was performed on DNA isolated from peripheral blood or bone marrow samples at diagnosis. Analysis was performed using the TruSight Myeloid Sequencing Panel (Illumina; San Diego, CA) on the MiSeq benchtop genome sequencer (Illumina). Of the 54 genes included in the panel, the complete coding region was sequenced in 15, with hotspot region coverage provided for 39.
Demographic and clinical data were obtained from the Princess Margaret Cancer Centre Registry.
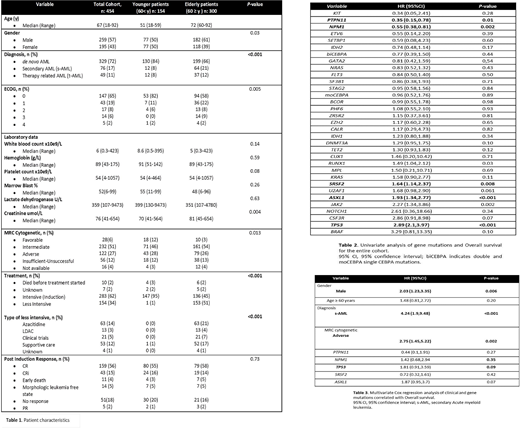
We identified 454 patients with a new diagnosis of AML in whom frontline NGS was performed. Of these, 300 were 60 years (range 60-92) or older. Demographic and clinical data are shown in Table 1. The median age overall at presentation was 67 yrs (range 18-92) and 57% were male. de novo AML was diagnosed in 329 patients (72%), secondary AML in 17%, and therapy related myeloid neoplasm in 11%. Secondary AML (sAML) was more frequently seen in the ≥60 vs the <60 population (21% vs 8%). Cytogenetic risk stratification by MRC was favorable in 6%, intermediate in 51%, and adverse in 27%. Unsuccessful cytogenetic testing occurred in 12%.
In total 283 patients (62%) received intensive chemotherapy (95% in the younger group and 45% in the older cohort). 216 patients (76%) achieved a response (CR/CRi/morphologic leukemia free state). No statistically significant difference was seen between age groups (P=0.73)
NGS detected 1655 variants. Of these, 1353 classified as oncogenic and 302 as variants of unknown significance (the latter were excluded from subsequent analysis). 14 patients had no mutations identified by NGS.
Forty three genes were recurrently mutated in the study cohort: 36 were mutated in >1% of patients, and 11 genes were mutated in >10% of patients. The most commonly mutated genes included DNMT3A (25%), NPM1 (21%), TET2 (19%), RUNX1 (16%), TP53 (16%), ASXL1 (15%), IDH2 (15%), SRSF2 (15%), NRAS (12%), and IDH1 (11%). Older patients had a greater number of mutations overall compared to younger adults (median: 3 vs 2; P = 0.001). In total 986 oncogenic variants were identified in the older group (median 3, range 1-12), while 367 oncogenic variants (median 0, range 0-2) were present in patients < 60 years old (P < 0.001). The older group more commonly harbored mutations in TET2 (25 vs 10%), ASXL1 (21 vs 4%), RUNX1 (20 vs 8 %), SRFS2 (20 vs 6 %), STAG2 (11 vs 6%) and U2AF1 (8 vs 2%).
Patients with PTPN11 and NPM1 mutations had longer OS, while patients carrying mutations in ASXL1, JAK2, RUNX1, TP53 and SRSF2 had a shorter OS (Table 2). No differences in OS between age groups (P= 0.2). Multivariate Cox regression analysis showed that male sex (P= 0.0057), sAML (P<0.001), MRC adverse cytogenetic analysis (P<0.002), ASXL1 mutations (P= 0.071) and TP53 mutations (P= 0.089), were associated with shorter OS (Table 3).
We then analyzed the correlation between mutations and response rate in patients receiving intensive treatment. PTPN11 and NPM1 mutations were associated with a greater likelihood of achieving a response, while patients with mutations in BCOR, TP53, SF3B1 and U2AF1 had a lower chance of response. Multivariate Cox regression analysis of mutations known to affect response did not show any differences by age, gender, or diagnosis. Mutations in BCOR (P= 0.045), TP53 (P= 0.032) and U2FA1 (P=0.019) were significantly associated with primary induction failure.
Overall, older AML patients harbored a greater number of mutations than did their younger counterparts. No age-related differences were observed in mutations known to affect response and/or OS. TP53 mutations, adverse cytogenetics and sAML, were poor prognostic factors regardless of age.
Schimmer:Medivir AB: Research Funding; Novartis: Consultancy, Membership on an entity's Board of Directors or advisory committees; Jazz Pharmaceuticals: Consultancy; Otsuka Pharmaceuticals: Consultancy. Yee:Agensys, Astex, GSK, Onconova, Genentech/Roche: Research Funding; Celgene, Novartis, Otsuka: Membership on an entity's Board of Directors or advisory committees. Gupta:Novartis: Consultancy, Honoraria, Research Funding; Incyte: Research Funding. Maze:Novartis: Consultancy, Honoraria. Schuh:Novartis: Consultancy; Otsuka: Consultancy; Pfizer: Consultancy; Teva: Consultancy; Celgene: Consultancy; Amgen Inc.: Consultancy; Shire: Consultancy; Jazz: Consultancy.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal